Evo Devo
Evolutionary development - at the biological interface between genetic regulatory mechanisms and biological evolution.
Evolutionary developmental biology (informally, evo-devo) is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved.
Evo Devo
A short form for 'evolutionary development', Evo Devo is a branch of biology that addresses the interface between evolution and development of individuals (ontogeny).
This site will examine genetic regulatory mechanisms that operate during embryologic development, and which have evolved through time to amplify the phenotypic manifestations of genotypic evolution.
"Once seen as distinct, yet complementary disciplines, developmental biology and evolutionary studies have recently merged into an exciting and fruitful relationship. The official union occurred in 1999 when evolutionary developmental biology, or "evo-devo," was granted its own division in the Society for Integrative and Comparative Biology (SICB). It was natural for evolutionary biologists and developmental biologists to find common ground. Evolutionary biologists seek to understand how organisms evolve and change their shape and form. The roots of these changes are found in the developmental mechanisms that control body shape and form. Developmental biologists try to understand how alterations in gene expression and function lead to changes in body shape and pattern. So although SICB only recently validated evo-devo as an independent research area, evo-devo really started over a decade ago when biologists began using an individual organism's developmental gene expression patterns to explain how groups of organisms evolved." Corey S. Goodman, Bridget C. Coughlin The evolution of evo-devo biology PNAS April 25, 2000 Vol. 97, Issue 9, 4424-4425, April 25, 2000 [References]
Genetic expression has traditionally been understood as one gene-one protein. However, it has recently been established that 25,000 human genes can generate about 90,000 proteins. This versatility arises because much genetic expression – production of proteins – is regulated through reversible and transmissable epigenetic mechanisms, which act without an alteration of archival DNA. Alternative splicing, which permits the environment-sensitive, regulated production of multiple polypeptides and proteins from a single gene, increases responsiveness and complexity without a change in the genome. Primate-specific Alu elements, which act as retroposons continue to be replicated and reinserted into the genome, increasing the potential for novel protein combinations.
This site will examine genetic regulatory mechanisms that operate during embryologic development, and which have evolved through time to amplify the phenotypic manifestations of genotypic evolution.
"Once seen as distinct, yet complementary disciplines, developmental biology and evolutionary studies have recently merged into an exciting and fruitful relationship. The official union occurred in 1999 when evolutionary developmental biology, or "evo-devo," was granted its own division in the Society for Integrative and Comparative Biology (SICB). It was natural for evolutionary biologists and developmental biologists to find common ground. Evolutionary biologists seek to understand how organisms evolve and change their shape and form. The roots of these changes are found in the developmental mechanisms that control body shape and form. Developmental biologists try to understand how alterations in gene expression and function lead to changes in body shape and pattern. So although SICB only recently validated evo-devo as an independent research area, evo-devo really started over a decade ago when biologists began using an individual organism's developmental gene expression patterns to explain how groups of organisms evolved." Corey S. Goodman, Bridget C. Coughlin The evolution of evo-devo biology PNAS April 25, 2000 Vol. 97, Issue 9, 4424-4425, April 25, 2000 [References]
Genetic expression has traditionally been understood as one gene-one protein. However, it has recently been established that 25,000 human genes can generate about 90,000 proteins. This versatility arises because much genetic expression – production of proteins – is regulated through reversible and transmissable epigenetic mechanisms, which act without an alteration of archival DNA. Alternative splicing, which permits the environment-sensitive, regulated production of multiple polypeptides and proteins from a single gene, increases responsiveness and complexity without a change in the genome. Primate-specific Alu elements, which act as retroposons continue to be replicated and reinserted into the genome, increasing the potential for novel protein combinations.
alternative exons
Alternative exons are targets of alternative splicing.
Typical features of alternative exons:
1. on average are less than half the size of constitutive exons,
2. include weak 5' and/or 3' splice sites.
3. auxiliary elements aid or prevent the recognition of these exons by binding trans-acting factors depending upon cell type, developmental stage, disease state, or in different environments, and
4. frequency of inclusion of an alternative exon in the mRNA transcript depends on a balance between positive and negative regulation. Enhancer (+) and silencer (-) elements can be found within the alternative exon or the flanking introns (ESE, ISE, ESS, ISS).
Splicing regulation is controlled by multiple elements – for a particular alternative exon these can be different elements, multiple copies of the same element located at different sites, or a combination of both. Different sets of auxiliary elements regulate alternative exons, but those alternative exons that are regulated by the same trans-acting factors share some common elements.
Intronic elements can be distal, but are more often located in the introns adjacent to the alternative exon (exon-intron junction). In some cases, intronic elements overlap with, or are included within, the consensus splice site sequences that are recognized by the basal spliceosomal machinery.
Exon isoforms include:
a. extension/truncation of an exon,
b. cassette exons in which an exon is present in one transcript but absent in an isoform of the transcript,
c. alternating exons that are used in alternative transcripts in a mutually exclusive manner, and d. intron retention in which a nucleotide region is used as an exon in a transcript while it is an intron in an alternative transcript.
Cassette exon, alternating exon, and intron retention can be further characterised as 'complex' or 'simple' depending on whether the 5' or/and 3' flanking exons also undergo modifications. For example, the flanking exon may be extended or truncated, or the exon that flanks a retained intron may be cassetted or alternated.
Typical features of alternative exons:
1. on average are less than half the size of constitutive exons,
2. include weak 5' and/or 3' splice sites.
3. auxiliary elements aid or prevent the recognition of these exons by binding trans-acting factors depending upon cell type, developmental stage, disease state, or in different environments, and
4. frequency of inclusion of an alternative exon in the mRNA transcript depends on a balance between positive and negative regulation. Enhancer (+) and silencer (-) elements can be found within the alternative exon or the flanking introns (ESE, ISE, ESS, ISS).
Splicing regulation is controlled by multiple elements – for a particular alternative exon these can be different elements, multiple copies of the same element located at different sites, or a combination of both. Different sets of auxiliary elements regulate alternative exons, but those alternative exons that are regulated by the same trans-acting factors share some common elements.
Intronic elements can be distal, but are more often located in the introns adjacent to the alternative exon (exon-intron junction). In some cases, intronic elements overlap with, or are included within, the consensus splice site sequences that are recognized by the basal spliceosomal machinery.
Exon isoforms include:
a. extension/truncation of an exon,
b. cassette exons in which an exon is present in one transcript but absent in an isoform of the transcript,
c. alternating exons that are used in alternative transcripts in a mutually exclusive manner, and d. intron retention in which a nucleotide region is used as an exon in a transcript while it is an intron in an alternative transcript.
Cassette exon, alternating exon, and intron retention can be further characterised as 'complex' or 'simple' depending on whether the 5' or/and 3' flanking exons also undergo modifications. For example, the flanking exon may be extended or truncated, or the exon that flanks a retained intron may be cassetted or alternated.
Labels: alternative exons, cassette exons, consensus splice site sequences, ESE, ESS, intron retention, ISE, ISS, mRNA transcript, spliceosome, splicing regulation, weak splice sites
Alu elements
Alu elements are about 300 nucleotides in length and include a distinctive sequence that ends in a poly-A tail. The human gene's protein-generating capacity is considerably increased by the presence of Alu elements.
About 5 percent of alternatively spliced exons in the human genome contain an Alu sequence, probably resulting from insertion of an Alu element into an intron of a gene where it would normally be spliced out and so would not have any negative consequence for the primate.
Through mutation, however, an Alu segment can convert an intron into an exon by any alteration in the Alu sequence that generates a new 5' or 3' splice site within the intron, resulting in its recognition by the spliceosome as an exon. (Such mutations usually arise during replication.)
If the new Alu exon is only alternatively spliced-in, the organism can produce a new protein without losing the gene's original function. This results from the original, wild types of mRNA continuing to be synthesized when the Alu exon is spliced-out. Problems arise only when a mutated Alu becomes spliced constitutively such that the Alu exon is always spliced in to all the mRNAs transcripts, with the loss of the original protein.
A single nucleotide polymorphism (point mutation) is sufficient to convert some silent intronic Alu elements into real exons. At present, the human genome contains approximately 500,000 Alu elements located within introns, and 25,000 of those could become new exons by undergoing this single-point mutation. Thus, Alu sequences have the potential to continue to greatly enrich the stock of meaningful genetic information available for producing new human proteins.
Three genetic illnesses have so far been identified as being caused by misplaced Alu sequences: Alport and Sly syndromes and OAT deficiency.
(Sorek et al., Genome Research 2002; Lev-Maor et al., Science 2003; Sorek et al., Molecular Cell 2004). [s]
About 5 percent of alternatively spliced exons in the human genome contain an Alu sequence, probably resulting from insertion of an Alu element into an intron of a gene where it would normally be spliced out and so would not have any negative consequence for the primate.
Through mutation, however, an Alu segment can convert an intron into an exon by any alteration in the Alu sequence that generates a new 5' or 3' splice site within the intron, resulting in its recognition by the spliceosome as an exon. (Such mutations usually arise during replication.)
If the new Alu exon is only alternatively spliced-in, the organism can produce a new protein without losing the gene's original function. This results from the original, wild types of mRNA continuing to be synthesized when the Alu exon is spliced-out. Problems arise only when a mutated Alu becomes spliced constitutively such that the Alu exon is always spliced in to all the mRNAs transcripts, with the loss of the original protein.
A single nucleotide polymorphism (point mutation) is sufficient to convert some silent intronic Alu elements into real exons. At present, the human genome contains approximately 500,000 Alu elements located within introns, and 25,000 of those could become new exons by undergoing this single-point mutation. Thus, Alu sequences have the potential to continue to greatly enrich the stock of meaningful genetic information available for producing new human proteins.
Three genetic illnesses have so far been identified as being caused by misplaced Alu sequences: Alport and Sly syndromes and OAT deficiency.
(Sorek et al., Genome Research 2002; Lev-Maor et al., Science 2003; Sorek et al., Molecular Cell 2004). [s]
alternative promoters
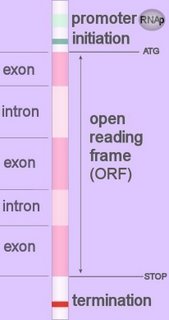 Promoters direct specialized enzymes to the location at which to commence reading the segment of DNA that codes for production of a protein.
Promoters direct specialized enzymes to the location at which to commence reading the segment of DNA that codes for production of a protein.Promoters can be divided into two broad categories: those without and those with CpG islands — stretches of DNA containing multiple copies of the nonmethlated dinucleotide CpG, which consists of the nucleotide cytosine (C) followed by guanine (G).
CpG islands comprise about 1 to 2% of the mammalian genome, and about 56% of sequenced human genes have CpG islands near their 5′ ends. This includes all genes that are ubiquitously expressed (housekeeping genes) plus many genes with a tissue-restricted pattern of expression. Promoters are normally located at the upstream edge of the CpG island, such that one or more of the 5′ exons of the gene generally fall within the island region. Although most CpG islands are nonmethylated in all tissues, a small proportion of islands become methylated during development [s].
In adults, single promoters with CpG islands tend to be linked to “housekeeping” genes, whereas single promoters without CpG islands are more often associated with highly regulated biological systems such as the immune and digestive systems.
A greater than expected percentage of mammalian genes have alternative promoters, which are more active during embryological development. Roughly 40-50 % of human and mouse genes have alternative promoters.
Alternative promoters can produce the same protein as single promoters, yet can be active in different tissues or at different times. In other cases, alternative promoters direct the polymerase enzymes to commence reading DNA at different start codons, ultimately resulting in different proteins with different functions.
Alternative promoters can affect gene expression in diverse ways. Production of different mRNA isoforms may be effected directly through different transcription start sites or indirectly through promoter-directed exon inclusion. The resulting transcripts may encode different protein isoforms, or they may vary only in their 5’ untranslated regions, affecting mRNA stability and translation efficiency.
Some genes employ promoters that differ in strength to direct tissue specific expression. Because tight regulation is essential for accurate gene function, loss of regulatory control can have serious disease-causing phenotypic effects such as malignancies resulting from activation of alternative promoters.
Alternative promoters, which confer greater flexibility, are more stable than single promoters over evolutionary time. Mammalian genomes are highly conserved and it is widely believed that gene regulation is largely responsible for the diversity of form and function between species. The higher evolutionary conservation of alternative promoters reflects the higher density of functional elements involved in regulating promoter choice.
Alternative promoters are tightly regulated, in line with their importance in cellular function. Cells with more than one promoter regulate which promoter to use, and when. Alternative promoters are more active during embryonic development.
Alternative Ways of Reading DNA Have Spurred Evolution : Green's studies of promoter function suggest intriguing hypotheses about evolutionary patterns. “The way C. elegans and a lot of other organisms increase their flexibility is simply to make a duplicate copy of a gene and have different regulation for the duplicate,” said Green. “With mammals, one way in which evolution has generated more diversity is to produce different versions of the same gene and allow the cell to regulate expression in multiple tissues. This is a way that evolution gets more bang for its buck, because it gets additional functions for the same gene.”
Labels: alternative promoter, conservation, CpG island, embryologic development, evolution, housekeeping genes, RNA polymerase
alternative splicing
Alternative splicing (AS) is a closely regulated, variable adaptation of the routine RNA modification process of pre-mRNA splicing. Alternative splicing is a form of epigenetic mechanism that enables a single gene to give rise to multiple, differentially spliced versions of a protein, increasing complexity without a change in the genome.
In 1980, a gene called IgM provided the first recognized example of alternative splicing in cells—there were earlier examples in viruses. It has since been demonstrated that cells employ alternative splicing to increase protein diversity toward a variety of biological ends.
An average mammalian gene possesses eight or nine exons—since most human genes undergoing some form of alternative splicing, virtually all of these exons are candidates for elaborate control. The human genome contains 3164.7 million nucleotide bases and only around 25,000 genes, much fewer than the 90,000 proteins that we are estimated to manufacture, and much fewer than prior estimates of 80,000-140,000 expressed sequence tags (ESTs). Human Genome Project
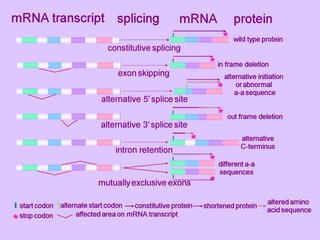 A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains, of a protein (ORF).
A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains, of a protein (ORF).
Alternative splicing can alter the mRNA product in several ways. (click to enlarge image at left)
At the simplest level, an exon can be removed (exon skip), lengthened or shortened (alternative 5'AS or 3'AS splicing). Thus, observed mechanisms of alternative splicing alteration include exon skipping, intron retention, and the use of an alternative splice donor or acceptor site. [r]
Alternative splicing can affect alternative promoters, internal exons, or alternative terminal exons, and generates segments of mRNA variability that shift the reading frame, insert or remove amino acids, or introduce a premature termination codon (Fig. 2). The variable segment within the mRNA results from insertion/deletion, or a mutually exclusive swap. The effects on coding potential are an in-frame insertion or deletion, a reading-frame shift, or introduction of a stop codon. mRNAs containing a stop codon >50 nt upstream of the position of the terminal intron are degraded by nonsense-mediated decay. Therefore, introduction of a premature termination codon into an mRNA by alternative splicing can be a mechanism to down-regulate expression of a gene. Alternative splicing also affects gene expression by removing or inserting regulatory elements controlling mRNA stability (NMD or nonstop decay), translation, or localization.
Exon skipping is the most frequent alternative splicing mechanism known in mammals, constituting about 40% of AS events, and as such is a major contributor to mammalian protein diversity. Exon skipping results in the loss of an exon in the alternatively spliced mRNA.
Alternative donor 5'AS or alternative acceptor 3'AS splicing together contribute to 25% of all AS events conserved between humans and mice (4).
Intron retention is defined by the presence of a transcript-confirmed intron within a transcript-confirmed exon. Intron retention occurs when introns are not spliced out of the RNA transcript, resulting in the intron(s) being retained within the mRNA as part of an exon. This lengthening mechanism the commonest form of alternative splicing in plants and lower multicellular organisms.
Regulation of AS:
Alternative splicing depends upon a splice-site and nearby enhancer and repressor sequences—short segments of RNA that couple with regulatory proteins. It has been estimated that the splicing of a single exon may be promoted by at least three to seven enhancer sequences.
As a result of alternate splicing, mutations that alter a splice site or a nearby regulatory sequence can have subtle effects by shifting the ratio of the resulting proteins without entirely eliminating any form.
Regulation of alternative splicing is often temporal or tissue-specific, generating different protein isoforms in different developmental states or different tissues. At the level of the organism, specific isoforms are known to be produced as a consequence of regulation by extracellular signaling mechanisms. Alternative splicing can allow one gene to generate different proteins in different tissues, providing the diversity encoded within genomes that are smaller than would be expected from gene expression by the organism. Many highly specialized brain proteins arise from differential splicing of genes that are also expressed in other tissues. Cells can even modify splicing in response to changing conditions, and not only can alternative splicing tweak the structure of a single protein, but it may also be a means of regulating entire pathways.
Recent bioinformatics studies have demonstrated that AS-prone exons can be distinguished from constitutively spliced exons by several features(1).
1. Divisibility by three of the exon length, which is likely to ensure the preservation of the reading frame in the mRNA (3).
2. Evolutionary conservation of intronic sequences flanking AS exons (3–8). Though not yet fully understood, the unusually high conservation of the introns that surround AS exons suggests the presence of cis-regulatory elements with regulatory proteins (5).
Alternative splicing is controlled by the binding of trans-acting protein factors to cis-acting sequences within the pre-mRNA, which result in differential use of splice sites. Since the splice sites do not contain enough information to explain the complex regulation of AS, this fine-tuned mechanism is achieved by multiple weak signals across the exons and introns which are recognized by an extensive number of different proteins (2). Many such alternative-binding sequences have been identified and are grouped as either enhancer or suppressor elements. These elements are typically short (8-10 nucleotides) and are less conserved than the splice sites at exon-intron junctions. The elements are either intronic or exonic, and either enhancers or silencers depending upon whether they increase or decrease splicing – ISE, ISS, ESE, ESS.
Members of the SR (serine rich) splicing factors control alternative splice site recognition by binding to the splicing enhancer and inhibitor elements (ESE, ESS, ISE, ISS). The SR proteins interact with the pre-mRNA substrate and with snRNP proteins (small nuclear ribonucleoproteins , pronounced 'snurps'). The role of SR splicing factors in regulating splice site selection is believed to occur in arginine-serine (RS) domain dependent and RS domain independent mechanisms.
animation of alternative splicing if this link does not work then click on figure 1 of alternative splicing link : life cycle of an mRNA ~ click on Quicktime Q :HHMI Feature Article on Alternative Splicing : Artist's conception of ASControlling the Synapse — 49 Proteins at a Time : The Alternative Splicing Website : Alternative Splicing DB (ASDB) : DNA-RNA-ProteinNational Center for Biotechnology Information
In 1980, a gene called IgM provided the first recognized example of alternative splicing in cells—there were earlier examples in viruses. It has since been demonstrated that cells employ alternative splicing to increase protein diversity toward a variety of biological ends.
An average mammalian gene possesses eight or nine exons—since most human genes undergoing some form of alternative splicing, virtually all of these exons are candidates for elaborate control. The human genome contains 3164.7 million nucleotide bases and only around 25,000 genes, much fewer than the 90,000 proteins that we are estimated to manufacture, and much fewer than prior estimates of 80,000-140,000 expressed sequence tags (ESTs). Human Genome Project
 A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains, of a protein (ORF).
A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains, of a protein (ORF).Alternative splicing can alter the mRNA product in several ways. (click to enlarge image at left)
At the simplest level, an exon can be removed (exon skip), lengthened or shortened (alternative 5'AS or 3'AS splicing). Thus, observed mechanisms of alternative splicing alteration include exon skipping, intron retention, and the use of an alternative splice donor or acceptor site. [r]
Alternative splicing can affect alternative promoters, internal exons, or alternative terminal exons, and generates segments of mRNA variability that shift the reading frame, insert or remove amino acids, or introduce a premature termination codon (Fig. 2). The variable segment within the mRNA results from insertion/deletion, or a mutually exclusive swap. The effects on coding potential are an in-frame insertion or deletion, a reading-frame shift, or introduction of a stop codon. mRNAs containing a stop codon >50 nt upstream of the position of the terminal intron are degraded by nonsense-mediated decay. Therefore, introduction of a premature termination codon into an mRNA by alternative splicing can be a mechanism to down-regulate expression of a gene. Alternative splicing also affects gene expression by removing or inserting regulatory elements controlling mRNA stability (NMD or nonstop decay), translation, or localization.
Exon skipping is the most frequent alternative splicing mechanism known in mammals, constituting about 40% of AS events, and as such is a major contributor to mammalian protein diversity. Exon skipping results in the loss of an exon in the alternatively spliced mRNA.
Alternative donor 5'AS or alternative acceptor 3'AS splicing together contribute to 25% of all AS events conserved between humans and mice (4).
Intron retention is defined by the presence of a transcript-confirmed intron within a transcript-confirmed exon. Intron retention occurs when introns are not spliced out of the RNA transcript, resulting in the intron(s) being retained within the mRNA as part of an exon. This lengthening mechanism the commonest form of alternative splicing in plants and lower multicellular organisms.
Regulation of AS:
Alternative splicing depends upon a splice-site and nearby enhancer and repressor sequences—short segments of RNA that couple with regulatory proteins. It has been estimated that the splicing of a single exon may be promoted by at least three to seven enhancer sequences.
As a result of alternate splicing, mutations that alter a splice site or a nearby regulatory sequence can have subtle effects by shifting the ratio of the resulting proteins without entirely eliminating any form.
Regulation of alternative splicing is often temporal or tissue-specific, generating different protein isoforms in different developmental states or different tissues. At the level of the organism, specific isoforms are known to be produced as a consequence of regulation by extracellular signaling mechanisms. Alternative splicing can allow one gene to generate different proteins in different tissues, providing the diversity encoded within genomes that are smaller than would be expected from gene expression by the organism. Many highly specialized brain proteins arise from differential splicing of genes that are also expressed in other tissues. Cells can even modify splicing in response to changing conditions, and not only can alternative splicing tweak the structure of a single protein, but it may also be a means of regulating entire pathways.
Recent bioinformatics studies have demonstrated that AS-prone exons can be distinguished from constitutively spliced exons by several features(1).
1. Divisibility by three of the exon length, which is likely to ensure the preservation of the reading frame in the mRNA (3).
2. Evolutionary conservation of intronic sequences flanking AS exons (3–8). Though not yet fully understood, the unusually high conservation of the introns that surround AS exons suggests the presence of cis-regulatory elements with regulatory proteins (5).
Alternative splicing is controlled by the binding of trans-acting protein factors to cis-acting sequences within the pre-mRNA, which result in differential use of splice sites. Since the splice sites do not contain enough information to explain the complex regulation of AS, this fine-tuned mechanism is achieved by multiple weak signals across the exons and introns which are recognized by an extensive number of different proteins (2). Many such alternative-binding sequences have been identified and are grouped as either enhancer or suppressor elements. These elements are typically short (8-10 nucleotides) and are less conserved than the splice sites at exon-intron junctions. The elements are either intronic or exonic, and either enhancers or silencers depending upon whether they increase or decrease splicing – ISE, ISS, ESE, ESS.
Members of the SR (serine rich) splicing factors control alternative splice site recognition by binding to the splicing enhancer and inhibitor elements (ESE, ESS, ISE, ISS). The SR proteins interact with the pre-mRNA substrate and with snRNP proteins (small nuclear ribonucleoproteins , pronounced 'snurps'). The role of SR splicing factors in regulating splice site selection is believed to occur in arginine-serine (RS) domain dependent and RS domain independent mechanisms.
animation of alternative splicing if this link does not work then click on figure 1 of alternative splicing link : life cycle of an mRNA ~ click on Quicktime Q :HHMI Feature Article on Alternative Splicing : Artist's conception of ASControlling the Synapse — 49 Proteins at a Time : The Alternative Splicing Website : Alternative Splicing DB (ASDB) : DNA-RNA-ProteinNational Center for Biotechnology Information
alternative 3' splicing
AS can also be attained by altering the position of the splice acceptor, alternative 3' splice site (3'AS). Together with alternative donor 5' splice site (5'AS), 3'AS contributes 25% of all AS events conserved between humans and mice (4).
"Recently, Hiller et al. (9) reported a widespread occurrence of a NAGNAG 3' acceptor splice site motif in the human genome. The NAGNAG motif includes two 3' splice site motifs in tandem and thus has the potential of producing mRNA isoforms which differ by a 3 nt sequence (NAG). Based on their analysis, Hiller et al. (9) suggested that the NAGNAG motif, which can insert or delete a single amino acid in the protein, is present in 30% of human genes and is functional in at least 5% of the genes. In addition it has been observed that alternative spliced isoforms, resulting from the NAGNAG motif are differentially expressed in human and mouse tissues (9,10).
...findings suggest that the selection of an acceptor site (3'AS) depends on the sequence environment, and can be altered by subtle changes such as point mutations. This is consistent with other studies showing that the pattern of AS can be altered by mutations in exonic splicing enhancer sites (ESEs) and exonic splicing silencer [sic] sites (ESSs) (16).
... by itself, the NAGNAG motif is not sufficient for AS. Nevertheless, analysis of a subset of the NAGNAG sites confirmed by expressed sequence tag (EST) data to be alternatively spliced shows that they encompass several characteristics of other known AS events. Comparison between the constitutively and alternatively spliced NAGNAG sites revealed that they differ principally by three major properties: (i) the sequence and evolutionary conservation of the NAGNAG motif, (ii) the conservation of intron sequences flanking the NAGNAG site, and (iii) the abundance of known cis-regulatory elements in the neighboring regions of the 3' splice sites. " [Alternative splicing regulation at tandem 3' splice sites]
Regulation of alternative 3' splice site selection by constitutive splicing factors.
Polypyrimidine tract binding protein (PTB) was found to inhibit the splicing of introns containing a strong binding site for this factor. However, the inhibitory effect of PTB could be partially reversed if pre-mRNAs were preincubated with U2 auxiliary factor (U2AF) prior to splicing in PTB-supplemented extracts. For alpha-tropomyosin, regulation of splicing by PTB and U2AF primarily affected the joining of exons 1-3 with no dramatic increases in 1-2 splicing being detected. Preincubation of pre-mRNAs with SR proteins led to small increases in 1-2 splicing. However, if pre-mRNAs were preincubated with SR proteins followed by splicing in PTB-supplemented extracts, there was a nearly complete reversal of the normal 1-2 to 1-3 splicing ratios. Thus, multiple pairwise, and sometimes antagonizing, interactions between constitutive pre-mRNA splicing factors and the pre-mRNA can regulate 3' splice site selection.
Lin CH, Patton JG. Regulation of alternative 3' splice site selection by constitutive splicing factors. RNA. 1995 May;1(3):234-45.
"Recently, Hiller et al. (9) reported a widespread occurrence of a NAGNAG 3' acceptor splice site motif in the human genome. The NAGNAG motif includes two 3' splice site motifs in tandem and thus has the potential of producing mRNA isoforms which differ by a 3 nt sequence (NAG). Based on their analysis, Hiller et al. (9) suggested that the NAGNAG motif, which can insert or delete a single amino acid in the protein, is present in 30% of human genes and is functional in at least 5% of the genes. In addition it has been observed that alternative spliced isoforms, resulting from the NAGNAG motif are differentially expressed in human and mouse tissues (9,10).
...findings suggest that the selection of an acceptor site (3'AS) depends on the sequence environment, and can be altered by subtle changes such as point mutations. This is consistent with other studies showing that the pattern of AS can be altered by mutations in exonic splicing enhancer sites (ESEs) and exonic splicing silencer [sic] sites (ESSs) (16).
... by itself, the NAGNAG motif is not sufficient for AS. Nevertheless, analysis of a subset of the NAGNAG sites confirmed by expressed sequence tag (EST) data to be alternatively spliced shows that they encompass several characteristics of other known AS events. Comparison between the constitutively and alternatively spliced NAGNAG sites revealed that they differ principally by three major properties: (i) the sequence and evolutionary conservation of the NAGNAG motif, (ii) the conservation of intron sequences flanking the NAGNAG site, and (iii) the abundance of known cis-regulatory elements in the neighboring regions of the 3' splice sites. " [Alternative splicing regulation at tandem 3' splice sites]
Regulation of alternative 3' splice site selection by constitutive splicing factors.
Polypyrimidine tract binding protein (PTB) was found to inhibit the splicing of introns containing a strong binding site for this factor. However, the inhibitory effect of PTB could be partially reversed if pre-mRNAs were preincubated with U2 auxiliary factor (U2AF) prior to splicing in PTB-supplemented extracts. For alpha-tropomyosin, regulation of splicing by PTB and U2AF primarily affected the joining of exons 1-3 with no dramatic increases in 1-2 splicing being detected. Preincubation of pre-mRNAs with SR proteins led to small increases in 1-2 splicing. However, if pre-mRNAs were preincubated with SR proteins followed by splicing in PTB-supplemented extracts, there was a nearly complete reversal of the normal 1-2 to 1-3 splicing ratios. Thus, multiple pairwise, and sometimes antagonizing, interactions between constitutive pre-mRNA splicing factors and the pre-mRNA can regulate 3' splice site selection.
Lin CH, Patton JG. Regulation of alternative 3' splice site selection by constitutive splicing factors. RNA. 1995 May;1(3):234-45.
alternative 5' splicing
The alternative donor 5' site operates in or other of two mechanisms to alter the protein product. Alternative splicing of internal exons can generate cassette, alternative 5' splice sites, alternative 3' splice sites, intron retention, and mutually exclusive splicing.
Due to alternative promoters, selection of one of several possible first exons results in variability at the 5' terminus of the mRNA. The determinative regulatory step in this mechanism is operation of a promoter rather than splice-site selection. The effect on the coding potential depends on the location of the translation initiation codon:
1. If translation initiates close to the first exons, the encoded proteins will contain alternative N termini.
2. If translation initiates in the common exon, different mRNAs will be generated that contain different 5' untranslated regions but that encode identical proteins.
Variation in alternative splicing across human tissues.
Controlling for differences in EST coverage among tissues, we found that the brain and testis had the highest levels of exon skipping. The most pronounced differences between tissues were seen for the frequencies of alternative 3' splice site and alternative 5' splice site usage, which were about 50 to 100% higher in the liver than in any other human tissue studied. Quantifying differences in splice junction usage, the brain, pancreas, liver and the peripheral nervous system had the most distinctive patterns of AS. Analysis of available microarray expression data showed that the liver had the most divergent pattern of expression of serine-arginine protein and heterogeneous ribonucleoprotein genes compared to the other human tissues studied, possibly contributing to the unusually high frequency of alternative splice site usage seen in liver. Sequence motifs enriched in alternative exons in genes expressed in the brain, testis and liver suggest specific splicing factors that may be important in AS regulation in these tissues.
Gene Yeo, Dirk Holste, Gabriel Kreiman and Christopher B Burge Free Full Text Variation in alternative splicing across human tissues. Genome Biology 2004, 5:R74
Due to alternative promoters, selection of one of several possible first exons results in variability at the 5' terminus of the mRNA. The determinative regulatory step in this mechanism is operation of a promoter rather than splice-site selection. The effect on the coding potential depends on the location of the translation initiation codon:
1. If translation initiates close to the first exons, the encoded proteins will contain alternative N termini.
2. If translation initiates in the common exon, different mRNAs will be generated that contain different 5' untranslated regions but that encode identical proteins.
Variation in alternative splicing across human tissues.
Controlling for differences in EST coverage among tissues, we found that the brain and testis had the highest levels of exon skipping. The most pronounced differences between tissues were seen for the frequencies of alternative 3' splice site and alternative 5' splice site usage, which were about 50 to 100% higher in the liver than in any other human tissue studied. Quantifying differences in splice junction usage, the brain, pancreas, liver and the peripheral nervous system had the most distinctive patterns of AS. Analysis of available microarray expression data showed that the liver had the most divergent pattern of expression of serine-arginine protein and heterogeneous ribonucleoprotein genes compared to the other human tissues studied, possibly contributing to the unusually high frequency of alternative splice site usage seen in liver. Sequence motifs enriched in alternative exons in genes expressed in the brain, testis and liver suggest specific splicing factors that may be important in AS regulation in these tissues.
Gene Yeo, Dirk Holste, Gabriel Kreiman and Christopher B Burge Free Full Text Variation in alternative splicing across human tissues. Genome Biology 2004, 5:R74
cassette exons
A cassette exon is defined as an exon that is present in one mRNA transcript but absent in an isoform of the transcript.
1. Initial cassette exons are missing in one or more transcript. An initial exon is the 5' exon of a transcript. To be flagged as an initial cassette exon, the exon cannot occur as an internal exon in any transcript, that is, the initial cassette exon is the first exon in the transcript.
2. Terminal casette exons are the same as initial cassette exon, except that they occurs at the 3' end, that is, the terminal cassette exon is the final exon in the transcript.
3. Internal cassette exons are present as an internal exon in at least one transcript of the cluster, that is, neither first nor final. Internal cassette exons are presumed to be the most biologically relevant because truncated sequences may create artificial occurences of initital and terminal cassette exons.
1. Initial cassette exons are missing in one or more transcript. An initial exon is the 5' exon of a transcript. To be flagged as an initial cassette exon, the exon cannot occur as an internal exon in any transcript, that is, the initial cassette exon is the first exon in the transcript.
2. Terminal casette exons are the same as initial cassette exon, except that they occurs at the 3' end, that is, the terminal cassette exon is the final exon in the transcript.
3. Internal cassette exons are present as an internal exon in at least one transcript of the cluster, that is, neither first nor final. Internal cassette exons are presumed to be the most biologically relevant because truncated sequences may create artificial occurences of initital and terminal cassette exons.









































